We performed single nucleus RNA sequencing (snRNA-Seq) to characterize the epithelial and stromal compartments of IBD-derived and normal control Organ-Dish models—referred to as IBODs and NODs, respectively (Fig. 1). Unbiased clustering and marker-based annotation confirmed the presence of all major intestinal cell types—epithelial, stromal, and immune—in ODs, reflecting the cellular complexity of native tissue (Fig. 1a).
Notably, IBODs displayed distinct transcriptional profiles in the stromal compartment, consistent with the inflammatory and immunogenic features of IBD tissues. Compared to NODs, which showed limited immune representation (primarily cycling B cells), IBODs exhibited a broader immune repertoire, including cycling B cells, T cells, NK cells, and dendritic cells. This indicates an active immune microenvironment in IBODs, which is essential for modeling chronic inflammation and fibrosis.
We further analyzed differentially expressed genes in IBODs vs. NODs using gene ontology tools, GOrilla and REVIGO. The analyses revealed enrichment of pathways in IBODs associated with IL-1/IL-6 signaling, IFN-γ signaling, neutrophil degranulation, and chemokine-mediated recruitment signatures absent in NODs (Fig. 1b). Together, these findings confirm that IBODs faithfully recapitulate the cellular and molecular hallmarks of IBD. Their inclusion of epithelial, stromal, and immune compartments within a self-organizing structure enables modeling of complex cell–cell and cell–matrix interactions—critical for studying inflammation, fibrosis, and therapeutic response in a human-relevant system.
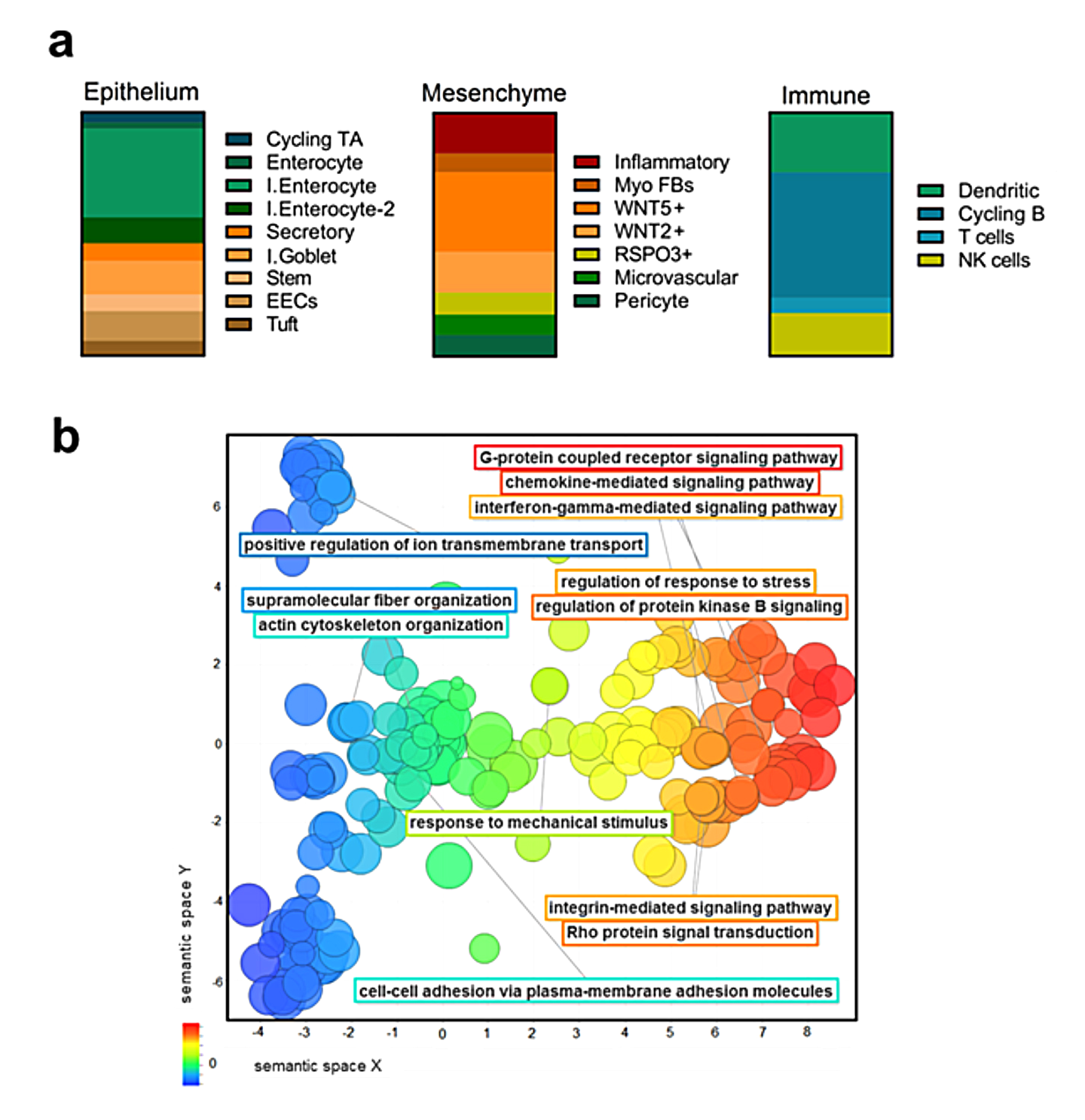
Figure 1. High-resolution transcriptomic analysis of IBODs reveals their unique parental signature.
(a) Single nucleus RNA-Seq analysis showing the presence of multiple cell types, including an immune cell signature exclusive to IBODs. Proportional plots illustrate subpopulations by cell type in IBODs. Cycling TA = cycling transient amplifying cells; I. enterocyte = immature enterocytes; I. goblet = immature goblet cells; Myo FB = myofibroblasts; cycling B = cycling B cells; NK = natural killer cells. (b) Enriched GO terms in IBODs vs. NODs, visualized as REVIGO scatterplots (P < 0.001). Terms are distributed along the X-axis based on semantic similarity of biological processes. Circle color indicates biological function; circle size reflects the number of genes associated with each term.
Although the precise pathogenesis of IBD remains unclear, epithelial wounding, reduced mucus secretion, and elevated levels of soluble inflammatory mediators secreted by stromal cells are believed to play key roles in the early development and progression of the disease [1, 2]. Notably, IBODs derived from patients with chronic IBD recapitulated both the histological and functional features of primary IBD tissues (Fig. 2). Hematoxylin and eosin (H&E) staining revealed distinct epithelial and stromal domains with a central lumen in both OD types (Fig. 2a). According to a board-certified pathologist, NODs predominantly displayed a simple columnar epithelium—characteristic of healthy colonic mucosa—whereas IBODs exhibited disorganized epithelium, including pseudostratified and stratified regions. These findings reflect the pseudo-stratification commonly observed in the colonic mucosa of patients with active IBD, typically arising from reparative epithelial responses to injury [3, 4]. Quantification of epithelial morphology across NODs, IBODs, and their respective primary tissues confirmed a significantly higher proportion of non-columnar epithelium in the IBD condition (Fig. 2b).
Immunohistochemical analysis of proliferation marker Ki67 revealed key distinctions between NODs and IBODs that mirror changes seen in matched patient tissues. In NODs, Ki67-positive cells were restricted to the base of a well-organized columnar epithelium, reflecting the proliferation gradient typical of healthy intestinal mucosa. By contrast, IBODs exhibited widespread Ki67 expression across disorganized, stratified epithelial layers (Fig. 2c) closely resembling colitic tissues from IBD patients [5, 6]. Quantification showed that over 80% of epithelial cells in IBODs were Ki67-positive, a proliferation rate consistent with regenerative epithelial turnover during active disease (Fig. 2d).

Figure 2. Disease-relevant features captured by
CaleoBio’s OD models.
IBODs reflect key histological and functional hallmarks of IBD, including epithelial disorganization, increased proliferation (Ki67), goblet cell depletion, and altered mucin secretion. These features align with matched IBD patient tissues, validating the capacity of OD models to mimic the in-vivo disease phenotype.
We performed single nucleus RNA sequencing (snRNA-Seq) to characterize the epithelial and stromal compartments of IBD-derived and normal control Organ-Dish models—referred to as IBODs and NODs, respectively (Fig. 1). Unbiased clustering and marker-based annotation confirmed the presence of all major intestinal cell types—epithelial, stromal, and immune—in ODs, reflecting the cellular complexity of native tissue (Fig. 1a).
Notably, IBODs displayed distinct transcriptional profiles in the stromal compartment, consistent with the inflammatory and immunogenic features of IBD tissues. Compared to NODs, which showed limited immune representation (primarily cycling B cells), IBODs exhibited a broader immune repertoire, including cycling B cells, T cells, NK cells, and dendritic cells. This indicates an active immune microenvironment in IBODs, which is essential for modeling chronic inflammation and fibrosis.
We further analyzed differentially expressed genes in IBODs vs. NODs using gene ontology tools, GOrilla and REVIGO. The analyses revealed enrichment of pathways in IBODs associated with IL-1/IL-6 signaling, IFN-γ signaling, neutrophil degranulation, and chemokine-
References
- Johansson ME, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. PNAS. 2008.
- Di Sabatino A, Ciccocioppo R, Morera R, et al. Role of the epithelium in IBD. World J Gastroenterol. 2012.
- Pai RK, Jairath V, Gutierrez A, et al. Histological criteria for the diagnosis of inflammatory bowel disease: a joint ECCO and ESP consensus statement. J Crohns Colitis. 2021.
- Wu XR, Lai Y, Chen L, et al. Histological features of ulcerative colitis and Crohn’s disease and their significance in diagnosis and disease assessment. Dig Med Res. 2022.
- Wong NA, Mayer NJ, MacKell S, Gilmour HM. Immunohistochemical assessment of Ki67 and p53 expression assists the diagnosis and grading of ulcerative colitis-related dysplasia. Histopathology. 2000.
- Clausen OP, Rognum TO, Brandtzaeg P. Ki-67: a useful marker for the evaluation of dysplasia in ulcerative colitis. J Clin Pathol. 1996.
- Dorofeyev AE, Vasilenko IV, Rassokhina OA, Kondratiuk RB. Mucosal barrier in ulcerative colitis and Crohn’s disease. Gastroenterol Res Pract. 2013.
- Johansson ME, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008